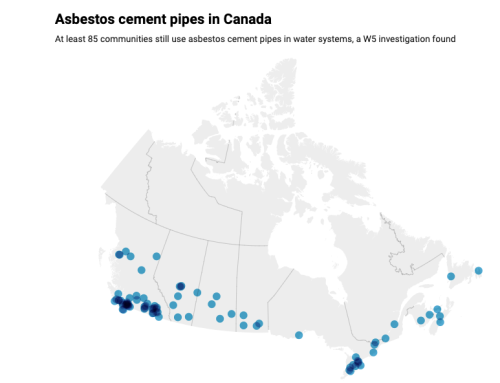
Hi all…with all the interest in, and potential promise of wastewater sampling for SARS-CoV-2 RNA it is important to understand that there are some obstacles which remain to be overcome. A just released article entitled “Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimisation and quality control are crucial for generating reliable public health information’ indicates that “in order to reliably interpret data produced from these efforts for informing public health interventions, additional quality control information and standardization in sampling design, sample processing, and data interpretation and reporting is needed.” About a dozen recommendations are identified to “enable robust interpretation of wastewater monitoring results and improved inferences regarding the relationship between monitoring results and disease cases.”
While preparing this, I noticed a similar article recently appeared in the same journal entitled “Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges.” The authors observe that “whilst being very promising, a number of areas were identified requiring additional research to further strengthen this approach and take full advantage of its potential. In particular, design of adequate sampling strategies, spatial and temporal resolution of sampling, sample storage, replicate sampling and analysis, controls for the molecular methods used for quantification of SARS-CoV-2 RNA in wastewater.”
Typically, I don’t draw attention to more than one article in my emails but given their similarity I felt it appropriate on this occasion. Note that while sometimes the links I provide are through Science Direct and may not be accessible to you, many COVID-19/SARS-CoV-2 articles are open access.
Bill
________________________________________________________
Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimisation and quality control are crucial for generating reliable public health information
Warish Ahmed, Aaron Bivins, Paul M. Bertsch, Kyle Bibby, Phil M. Choid Kata Farkase, Pradip Gyawalif , Kerry A. Hamilton, Eiji Haramoto, Masaaki Kitajima, Stuart L. Simpson, Sarmila Tandukar, Kevin Thomas, Jochen F. Mueller
Current Opinion in Environmental Science & Health Available online 30 September 2020
Abstract
“Monitoring for SARS-CoV-2 RNA in wastewater through the process of wastewater-based epidemiology (WBE) provides an additional surveillance tool, contributing to community-based screening and prevention efforts as these measurements have preceded disease cases in some instances. Numerous detections of SARS-CoV-2 RNA have been reported globally using various methods, demonstrating the technical feasibility of routine monitoring. However, in order to reliably interpret data produced from these efforts for informing public health interventions, additional quality control information and standardization in sampling design, sample processing, and data interpretation and reporting is needed. This review summarizes published studies of SARS-CoV-2 RNA detection in wastewater as well as available information regarding concentration, extraction, and detection methods. The review highlights areas for potential standardization including considerations related to sampling timing and frequency relative to peak fecal loading times; inclusion of appropriate information on sample volume collected; sample collection points; transport and storage conditions; sample concentration and processing; RNA extraction process and performance; effective volumes; PCR inhibition; process controls throughout sample collection and processing; PCR standard curve performance; and recovery efficiency testing.Researchers are recommended to follow the Minimum Information for Publication of Quantitative Real-Time PCR (MIQE) guidelines. Adhering to these recommendations will enable robust interpretation of wastewater monitoring results and improved inferences regarding the relationship between monitoring results and disease cases.”
_________________________________________________________
Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges
Gertjan Medema, Frederic Been, Leo Heijnen, Susan Petterson
Current Opinion in Environmental Science & Health DOI: https://doi.org/10.1016/j.coesh.2020.09.006
https://www.sciencedirect.com/science/journal/aip/24685844
https://www.sciencedirect.com/science/article/pii/S2468584420300635
Abstract
“Analysing wastewater can be used to track infectious disease agents that are shed via stool and urine. Sewage surveillance of SARS-CoV-2 has been suggested as tool to determine the extent of COVID-19 in cities and serve as early warning for (re-)emergence of SARS-CoV-2 circulation in communities. The focus of this review is on the strength of evidence, opportunities and challenges for the application of sewage surveillance to inform public health decision making. Considerations for undertaking sampling programs are reviewed including sampling sites, strategies, sample transport, storage and quantification methods; together with the approach and evidence base for quantifying prevalence of infection from measured wastewater concentration. Published SARS-CoV-2 sewage surveillance studies (11 peer reviewed, and 10 pre-prints) were reviewed to demonstrate the current status of implementation to support public health decisions. Whilst being very promising, a number of areas were identified requiring additional research to further strengthen this approach and take full advantage of its potential. In particular, design of adequate sampling strategies, spatial and temporal resolution of sampling, sample storage, replicate sampling and analysis, controls for the molecular methods used for quantification of SARS-CoV-2 RNA in wastewater. The use of appropriate prevalence data and methods to correlate or even translate SARS-CoV-2 concentrations in wastewater to prevalence of virus shedders in the population is discussed.”
Photo by Markus Spiske from Pexels